Course Description
This course introduces the mathematical modeling techniques needed to address key questions in modern biology. An overview of modeling techniques in molecular biology and genetics, cell biology and developmental biology is covered. Key experiments that validate mathematical models are also discussed, as well as …
This course introduces the mathematical modeling techniques needed to address key questions in modern biology. An overview of modeling techniques in molecular biology and genetics, cell biology and developmental biology is covered. Key experiments that validate mathematical models are also discussed, as well as molecular, cellular, and developmental systems biology, bacterial chemotaxis, genetic oscillators, control theory and genetic networks, and gradient sensing systems. Additional specific topics include: constructing and modeling of genetic networks, lambda phage as a genetic switch, synthetic genetic switches, circadian rhythms, reaction diffusion equations, local activation and global inhibition models, center finding networks, general pattern formation models, modeling cell-cell communication, quorum sensing, and finally, models for Drosophila development.
Course Info
Instructor
Departments
Learning Resource Types
notes
Lecture Notes
assignment
Problem Sets
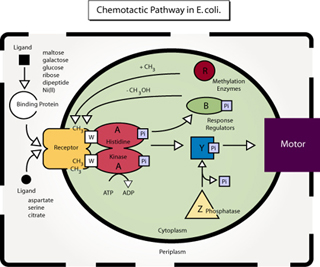
Diagram of the chemotactic pathway in E. coli. (Figure by MIT OCW. After figure 4 in Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz, and M. A. Danielson. “The Two-component Signaling Pathway of Bacterial Chemotaxis: A Molecular View of Signal Transduction by Receptors, Kinases, and Adaptation Enzymes.” In Annu Rev Cell Dev Biol. 13 (1997): 457-512.)








