Course Description
This course is an introduction to computational biology emphasizing the fundamentals of nucleic acid and protein sequence and structural analysis; it also includes an introduction to the analysis of complex biological systems. Topics covered in the course include principles and methods used for sequence alignment, …
This course is an introduction to computational biology emphasizing the fundamentals of nucleic acid and protein sequence and structural analysis; it also includes an introduction to the analysis of complex biological systems. Topics covered in the course include principles and methods used for sequence alignment, motif finding, structural modeling, structure prediction and network modeling, as well as currently emerging research areas.
Course Info
Learning Resource Types
theaters
Lecture Videos
notes
Lecture Notes
group_work
Projects
assignment_turned_in
Programming Assignments with Examples
assignment
Presentation Assignments
assignment
Written Assignments
Instructor Insights
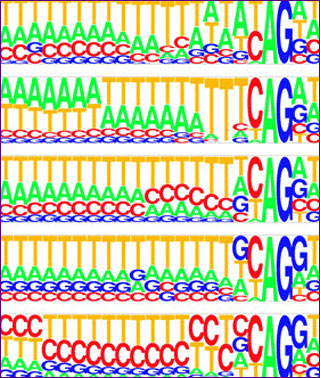
Splice signal motifs of five species, created using the PICTOGRAM program. The height of each letter is proportional to the frequency of the corresponding base at the given position, and bases are listed in descending order of frequency from top to bottom. Source: Figure 2 of Lim, Lee P., and Christopher B. Burge. “A Computational Analysis of Sequence Features Involved in Recognition of Short Introns.” Proceedings of the National Academy of Sciences 98, no. 20 (2001): 11193–8. (Courtesy of National Academy of Sciences)








