Course Meetings Times
Lectures: 2 sessions / week; 1.5 hours / session
Recitations: 1 session / week; 1 hour / session
Description
This course is an introduction to computational biology emphasizing the fundamentals of nucleic acid and protein sequence and structural analysis; it also includes an introduction to the analysis of complex biological systems. Topics covered in the course include principles and methods used for sequence alignment, motif finding, structural modeling, structure prediction and network modeling, as well as currently emerging research areas. This course is designed for advanced undergraduates and graduate students with strong backgrounds in either molecular biology or computer science, but not necessarily both. The scripting language Python—which is widely used for bioinformatics and computational biology—will be used; foundational material covering basic programming skills will be provided by the teaching assistants. Graduate versions of the course involve an additional project component.
Prerequisites
There are different prerequisites for the various versions of the course. See the table for clarification.
- 7.01 Fundamentals of Biology
- 7.05 General Biochemistry
- 5.07 Biological Chemistry
- 6.00 Introduction to Computer Science and Programming
- 6.01 Introduction to Electrical Engineering and Computer Science
- 18.440 Probability and Random Variables
- 6.041 Probabilistic Systems Analysis and Applied Probability
| DEPARTMENT |
COURSE NUMBER |
LEVEL | PREREQUISITES | RECITATIONS | ASSIGNMENTS | PROJECT | GRADING | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 7.01 | 7.05 or 5.07 | 6.00 or 6.01 | 18.440 or 6.041 | |||||||
| Biology | 7.36 | Undergraduate |

|

|

|
Optional; review of lectures | No | Grading scheme 1 | ||
| 7.91 | Graduate |

|

|

|
Optional; review of lectures | Yes | Grading scheme 2 | |||
| Biological Engineering | 20.390 | Undergraduate |
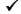
|
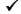
|
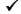
|
Optional; review of lectures | No | Grading scheme 1 | ||
| 20.490 | Graduate |
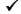
|
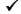
|
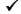
|
Optional; review of lectures | Yes | Grading scheme 2 | |||
| Electrical Engineering and Computer Science | 6.802 | Undergraduate |

|

|
Optional; review of lectures | No | Grading scheme 1 | |||
| 6.847 | Graduate |

|

|
Required; cover additional material related to artificial intelligence | Additional artificial intelligence problems will be assigned | Yes | Grading scheme 3 | |||
| Health Sciences and Technology | HST.506 | Graduate |
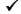
|
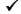
|
Optional; review of lectures | Yes | Grading scheme 2 | |||
Textbook
This textbook is recommended for the course:
Zvelebil, Marketa J., and Jeremy O. Baum. Understanding Bioinformatics. Garland Science, 2007. ISBN: 9780815340249. [Preview with Google books]
The instructors have also selected the following texts as particularly useful in specific areas, if you are looking for more information:
Background in Python Programming
Ascher, David, and Mark Lutz. Learning Python. 2nd ed. O’Reilly Media, Inc., 2003. ISBN: 9780596002817. [Preview with Google Books]
Martelli, Alex. Python in a Nutshell. O’Reilly Media. 2006. ISBN: 9780596100469. [Preview with Google Books]
Background in Molecular Biology, Cell Biology, and Biochemistry
Watson, James D., Tania A. Baker, et al. Molecular Biology of the Gene. Benjamin Cummings, 2013. ISBN: 9780321762436.
Alberts, Bruce, Alexander Johnson, et al. Molecular Biology of the Cell. Garland Science, 2007. ISBN: 9780815341055.
Berg, Jeremy M., John L. Tymoczko, et al. Biochemistry. W. H. Freeman, 2008. ISBN: 9781429235020.
Branden, Carl I., and John Tooze. Introduction to Protein Structure. Taylor & Francis, Inc., 2000.
Petsko, Gregory A., and Dagmar Ringe. Protein Structure and Function. Oxford University Press, 2008. ISBN: 9780199556847.
Other Useful References for the Class
Leach, Andrew. Molecular Modelling: Principles and Applications. Prentice Hall, 2001. ISBN: 9780582382107.
- An in-depth treatment of molecular mechanics and other modeling methods.
Alon, Uri. An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman and Hall / CRC, 2013. ISBN: 9781439837177. [Preview with Google Books]
- An introduction to topics related to biological networks from a physicist’s perspective.
Durbin, Richard, Sean R. Eddy, et al. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, 1998. ISBN: 9780521629713.
- Contains more in-depth discussion of many of the algorithms and methods discussed in class.
Glantz, Stanton. Primer of Biostatistics. McGraw-Hill Medical, 2011. ISBN: 9780071781503. [Preview with Google Books]
- Covers basic statistics with applications to biology and medicine.
Gonick, Larry, and Woollcott Smith. The Cartoon Guide to Statistics. Paw Prints, 2008. ISBN: 9781435242715.
- A very useful introduction or refresher. Fun, but generally accurate.
Assignments
Five written or computer-based problem sets will be assigned. These are designed to promote deeper understanding of the principles and algorithms discussed in class and to provide hands-on experience with bioinformatics tools. The scripting language Python—which is widely used for bioinformatics and computational biology—will be used in the problem sets. More information is in the Assignments section.
Exams
There will be two 80–minute exams. The material covered on each exam is non-cumulative; consult the Calendar to see which topics were covered on each exam. There is no final exam.
Project
Students in one of the graduate versions of this course will complete a computational biology research project. See the table for clarification. The project is designed to give you practice in applying computational methods to contemporary problems in biology. Students design and carry out projects working in a group or by themselves. All students will provide online feedback (peer review) on the presentations. More information is in the Project section.
Grading
The different versions of this course have different grading schemes. See the table for clarification. An additional 1% extra credit may be awarded for exceptional class participation.
Grading Scheme 1
| ACTIVITIES | PERCENTAGES |
|---|---|
| Problem Sets (out of 100 points) | 36 |
| Exams | 62 |
| Peer Review | 2 |
Grading Scheme 2
| ACTIVITIES | PERCENTAGES |
|---|---|
| Problem Sets (out of 100 points) | 30 |
| Exams | 48 |
| Project | 20 |
| Peer Review | 2 |
Grading Scheme 3
| ACTIVITIES | PERCENTAGES |
|---|---|
| Problem Sets (out of 100 points) | 25 |
| Extra Artificial Intelligence Problems | 5 |
| Exam | 48 |
| Project | 20 |
| Peer Review | 2 |










