Modules: 1.1 | 1.2 | 1.3 | 1.4 | 1.5 | 1.6 | 1.7
Introduction
Yesterday a few milliliters of LB+Kan broth were inoculated with some candidate colonies and the tubes were grown overnight at 37°C. Kanamycin was included in the broth to ensure that the cells would maintain the M13KO7 DNA. Today the cells will have grown to high density and the phage DNA will have undergone many replications. All the copies of the phage DNA in your overnight culture should be identical (“clones” of one another), since the culture began with a single colony and that colony grew from a single transformed cell. Next, you must isolate the plasmid DNA from the candidates and determine if any of the plasmids contain the modification you intended to construct.
To isolate the plasmid from the overnight cultures, you will perform what is commonly called a “mini-prep.” This term distinguishes the procedure from a “maxi-” or “large scale-prep” which involves a larger volume of cells and additional steps of purification. The overall goal of each “prep” is the same–to separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further.
In the traditional mini-prep protocol, which you will perform today, the media is removed from the cells by centrifugation. The cells are resuspended in “Solution I” which contains Tris to buffer the cells and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. A solution of sodium hydroxide and SDS is then added. The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. The pH of the solution is returned to neutral by the potassium acetate in “Solution III.” At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. In addition, the DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell.
Once you have isolated some plasmid DNA, you will perform some “diagnostic digests” to determine if any of the candidates have the desired construct. When choosing enzymes for diagnostic digests, it is good practice to choose enzymes that
- cut both the plasmid backbone and the insert.
- verify restriction sites on both sides of the insert.
- release a fragment that can be visualized on a gel. Fragments smaller than 500 basepairs are very hard to detect. Fragments larger than 7 kilobases are difficult to judge accurately on agarose gels, though differences between larger fragments are detectable.
- work best in the same buffer and at the same temperature.
This last rule is breakable but it is makes your labwork more complicated if you must change buffers or perform the reactions sequentially, at low then high temperatures.
While your gel is running, the class will discuss the journal article you read for today about refactoring T7. This class discussion will inform the M13 genome renovation you have started, and much of your assignment for next time will be to map your understanding of M13 phage biology to engineering efforts like the one described in the T7 paper.
Protocol
Sometime before you leave today you should count or estimate the number of colonies that arose from your transformation reactions. If the number of colonies is high, you can sector the back of the plate (using a Sharpie to draw sections that are 1/4 or 1/8th of the area) and count only the colonies in a sector, then multiplying to determine the total.
Part 1: Plasmid Miniprep
One of the teaching faculty has set up four overnight LB + kan cultures of bacteria for you to work with. If your reactions were unsuccessful then you have been provided with candidates from other reactions.
-
Label four eppendorf tubes (1, 2, 3 or 4). Vortex the bacteria then pour some into the appropriate eppendorf tube so that the tubes are almost full. If you are nervous about pouring the liquid, you can use your P1000 to pipet 750 µl into each eppendorf twice. Either way, the eppendorf should be quite full when you try to close the cap. You can wear gloves to keep the bacteria from splashing your skin or you can wash your hands after closing all the caps.
-
Return any remaining volume of cells to the teaching faculty, who will store them for you for next time.
-
Balance the eppendorf tubes in the microfuge, and then spin them for one minute.
-
Aspirate the supernatant, as shown, removing as few cells as possible.
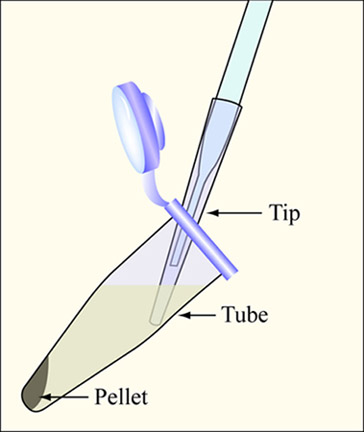
- Aspirate the supernatant, as shown, removing as few cells as possible. Make sure to use a yellow pipet tip on the aspirator and change between samples. (Figure by MIT OpenCourseWare.)
- Resuspend the cells in 100 µl of Solution I, changing tips between samples.
- Prepare Solution II by mixing 500 µl of 2% SDS with 500 µl of 0.4M NaOH in an eppendorf tube. Add 200 µl of Solution II to each sample and invert the tubes five or six times to mix. In some cases the samples may appear to “clear” but don’t worry if you don’t see a big change. Place the tubes on ice for five minutes.
- Add 150 µl of Solution III to each tube and immediately vortex each tube for 10 seconds with your vortex set at the highest setting. White clumps should appear in the solution after you vortex it. Place the tubes in the room temperature microfuge and spin them for 4 minutes.
- While the tubes are spinning, label another set of eppendorf tubes with the plasmid names and your team color.
- A white pellet should be visible when you remove your tubes from the microfuge. Use your P1000 to transfer 400 µl of each supernatant to the appropriate clean eppendorf tube. It’s OK to leave some of the supernatant behind. Avoid transferring any of the white pellet.
- Add 1000 µl of room temperature 100% ethanol to each new tube. The tubes will be quite full. Close the caps and invert the tubes at least five times to thoroughly mix the contents.
- Microfuge the samples for 2 minutes. It is important to orient your tubes in the microfuge this time since the pellets from this spin will be barely visible.
- Remove the supernatants using your P1000 or the aspirator, but be careful not to disturb the pellet of plasmid DNA that is at the bottom of the tube. Remove as much of the supernatant as possible but you do not need to remove every drop since you will be washing the pellet in the next step.
- Add 500 µl of 70% ethanol to each pellet. Spin the samples one minute, orienting the tubes in the microfuge so you will know where to find the pellet. Immediately remove the supernatant with your P1000, making sure to keep the tip on the side of the tube that doesn’t have your pellet. Remove as much liquid as possible, using your P200 set to 100 µl, to remove the last few droplets.
- To completely dry the pellets, place your rack in the hood with the caps open for a few minutes. When the pellets are completely dry, add 50 µl of sterile water to each sample and vortex each tube for 2 X 30 seconds to completely dissolve the pellets. The liquid can be brought back to the bottom of the tubes by spinning them in the microfuge for a few seconds. Store the DNA on ice, and return any DNA you don’t use for Part 2 to the teaching faculty before you leave lab today (you’ll need it for the sequencing reactions next time!).
Part 2: Diagnostic Digests
You will perform two diagnostic digests on each of the plasmids you have prepared to see if any have the PCR insert. Use information from the lab manual, the NEB catalog and the plasmid maps you’ve drawn to choose the enzymes you’ll use. The following table may be helpful as you plan your work.
| DIGEST 1 | DIGEST 2 | |
|---|---|---|
| Plasmid DNA | 5 µl | 5 µl |
| 10X NEB buffer | 2.5 µl of buffer#_____ | 2.5 µl of buffer#_____ |
| Enzyme | 0.25 µl of _____ | 0.25 µl of _____ |
| 2nd Enzyme (if desired) | 0.25 µl of _____ | 0.25 µl of _____ |
| H2O | For a total volume of 25 µl | |
- Prepare two reaction cocktails with water, buffer and enzyme. Prepare enough of each cocktail for 5 digests. Leave the cocktails on ice.
- Aliquot 5 µl of plasmids into well labeled eppendorf tubes. The labels should include the plasmid name, the enzymes to be added and your team color.
- Add 20 µl of each cocktail to each tube. Flick the tubes to mix the contents then incubate the tubes at 37°C, for 30 minutes. You should have 8 digests total. When your samples are digesting, you should re-read the Refactoring T7 paper that we will discuss, focusing on one aspect of the article (figure 1, 2, 3 or 4, Supplementary Table 2, Supplementary Figure 1) that you will be assigned and that you will present to the class.
- Add 2 µl of loading dye to each of the digests you have assembled.
Part 3: Agarose Gel Electrophoresis
Load your samples on a 1% TAE agarose gel in the following order. Each group should fill 9 wells.
| LANES | SAMPLES | VOLUMES TO LOAD |
|---|---|---|
| 1 | 1 kb Marker | 5 µl |
| 2 | Candidate 1, Diagnostic digest 1 | All of reaction volume (~30µl) |
| 3 | Candidate 2, Diagnostic digest 1 | All of reaction volume (~30µl) |
| 4 | Candidate 3, Diagnostic digest 1 | All of reaction volume (~30µl) |
| 5 | Candidate 4, Diagnostic digest 1 | All of reaction volume (~30µl) |
| 6 | Empty | |
| 7 | Candidate 1, Diagnostic digest 2 | All of reaction volume (~30µl) |
| 8 | Candidate 2, Diagnostic digest 2 | All of reaction volume (~30µl) |
| 9 | Candidate 3, Diagnostic digest 2 | All of reaction volume (~30µl) |
| 10 | Candidate 4, Diagnostic digest 2 | All of reaction volume (~30µl) |
Once all the samples are loaded, the power will be applied (100V for 45 minutes) and the gel will be photographed. While you are waiting, the class will discuss the T7 paper.
DONE!
For Next Time
- Prepare a table with the results of your ligations and transformations. Calculate your transformation efficiency (# colonies/?g plasmid DNA) based on the transformation you performed with M13KO7. In three or four sentences, interpret the ligation results.
- Choose one of the following two essays to write a thoughtful response to their “fighting words.” Rebut the quoted statements by first explaining what the quote refers to, explaining why the author or quoted individual might have said it, and then provide at least five counter points or examples to support the opposite point of view. Draw your arguments from your experiments with M13 whenever possible. Print out two copies of this portion of the assignment. Next time you and your lab partner will exchange responses and provide feedback to each other on the writing and ideas within.
Essay 1: Choose one of the following quotes to address. Both come from Andrew Pollack in the New York Times, Tuesday, January 17, 2006, “Custom-Made Microbes, at Your Service” which quotes Professor Arnold of Caltech as saying:
- “(Synthetic Biology) has a catchy new name, but anybody over 40 will recognize it as good old genetic engineering applied to more complex problems.”
and
- “There is no such thing as a standard component, because even a standard component works differently depending on the environment. The expectation that you can type in a sequence and can predict what a circuit will do is far from reality and always will be.”
Essay 2: From “Editorial: Meanings of ’life’.” Nature 447 (2007): 1031-1032.
- “it would be a service…to dismiss the idea that life is a precise scientific concept”
Reagents List
- Solution I
- 25 mM Tris pH8
- 10 mM EDTA pH8
- 5 mM Glucose
- Solution II
- 1% SDS
- 0.2M NaOH
- Solution III
- 3M KAc, pH 4.8








