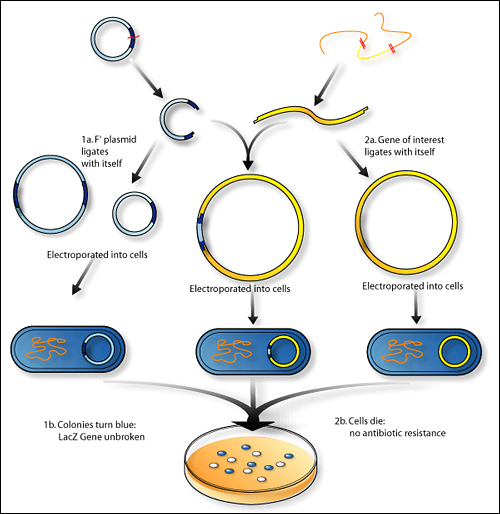
An illustration depicting cloning using a plasmid containing LacZ (image attributed to “The Science Creative Quarterly” and Jen Philpot).
This unit will cover some basic recombinant DNA technologies, why they were developed, and how they are used today in many different scientific arenas. We will introduce the different types of restriction enzymes, their advantages and disadvantages, and how they are used to create a recombinant DNA molecule. We will review how to make a recombinant genomic DNA library and how to use this library to find a specific gene. We will then outline how to use a library to clone a gene by complementation of a mutant phenotype. We will also review how to make a recombinant cDNA library and how to use this library to find a specific gene. In addition, we will outline the differences between a genomic and a cDNA library and discuss how to use a cDNA library to clone a gene of interest. Finally, we will discuss methods to analyze genes identified through recombinant DNA technologies.
During this unit, you will learn the steps involved in a basic cloning strategy. You will determine which restriction enzyme to use to create a desired piece of recombinant DNA, given specific DNA sequences. You will also learn the function of DNA ligase, understand how vectors are used, and learn how to construct a recombinant genomic DNA library. You will then demonstrate how to use a genomic library to identify a gene of interest, and how to clone the gene of interest by complementation of a mutant phenotype. Next, you will learn how a recombinant cDNA library is constructed, and how it is used to identify a gene of interest by the protein made. You will move on to learning about the uses of agarose gel electrophoresis to analyze DNA molecules, and learn the basic mechanism of DNA sequencing by the dideoxy chain termination method. Finally, you will learn about the polymerase chain reaction (PCR) method of amplifying DNA, and when to use this technique.
By the end of this unit, you will understand some basic techniques of recombinant DNA technologies, and some of their applications. You will understand the differences between the techniques you have learned about, as well as their uses and their limitations. Your knowledge of recombinant DNA technologies will culminate in your understanding of the importance of GenBank®, the vast database containing the DNA sequence of the entire genomes for many different organisms, and understanding why this is useful.
Looking for something specific in this course? The Resource Index compiles links to most course resources in a single page.








